Research
Highlighted
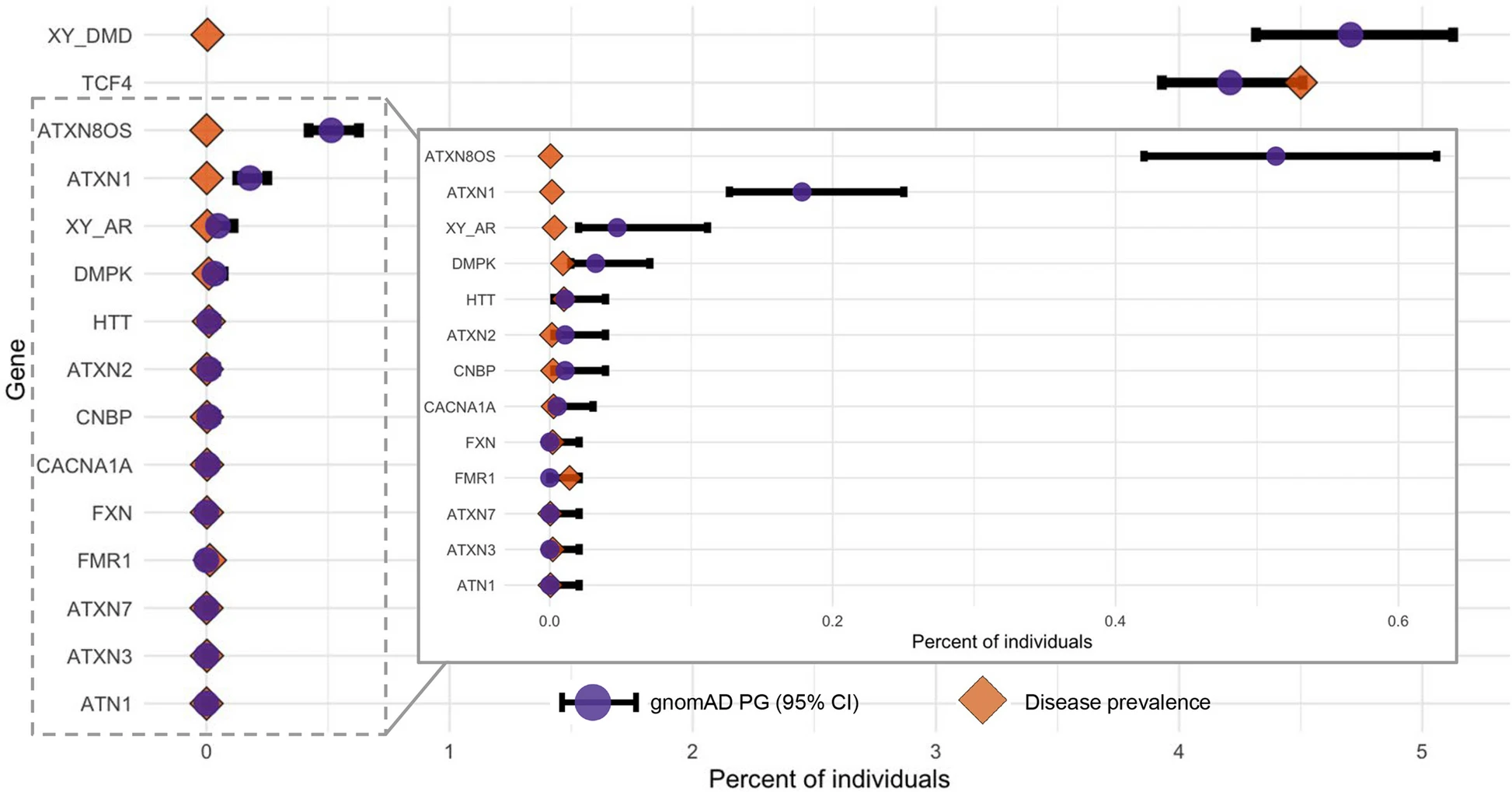
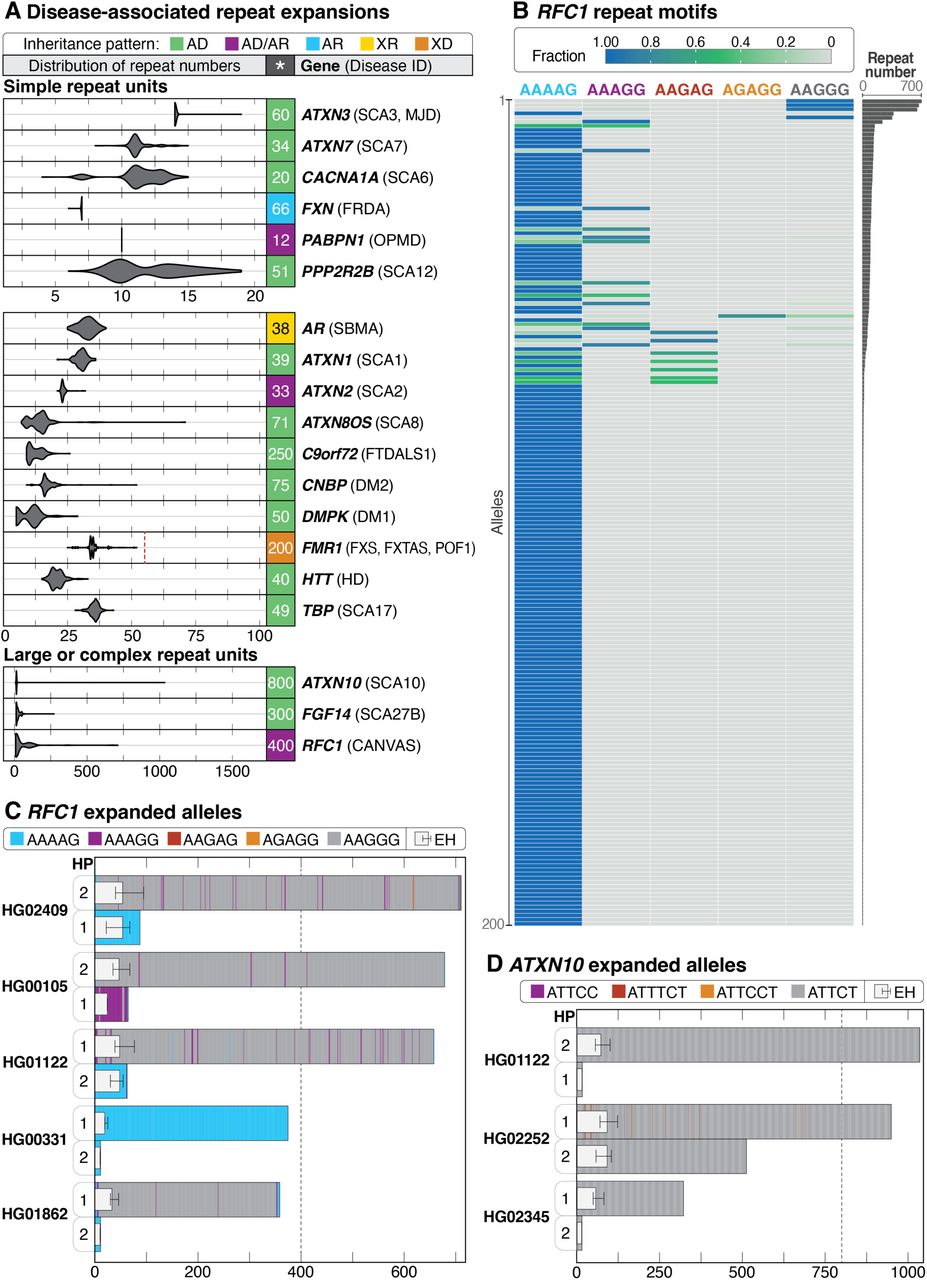
STRchive: a dynamic resource detailing population-level and locus-specific insights at tandem repeat disease loci
Genome Medicine
·
26 Mar 2025
·
doi:10.1186/s13073-025-01454-4
All
2025

The selective dynamics of interruptions at short tandem repeats
Cold Spring Harbor Laboratory
·
12 Jun 2025
·
doi:10.1101/2025.06.09.658724
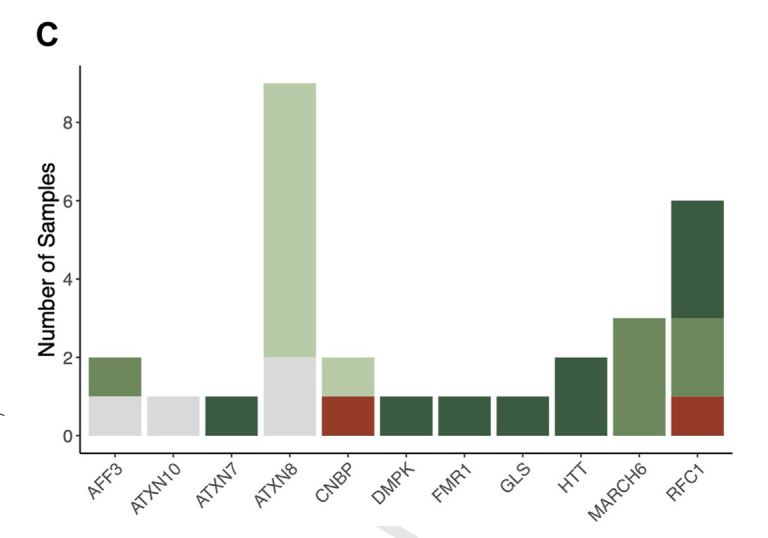
A genome-wide approach for the discovery of novel repeat expansion disorders in the Undiagnosed Diseases Network cohort
Genetics in Medicine
·
01 May 2025
·
doi:10.1016/j.gim.2025.101462
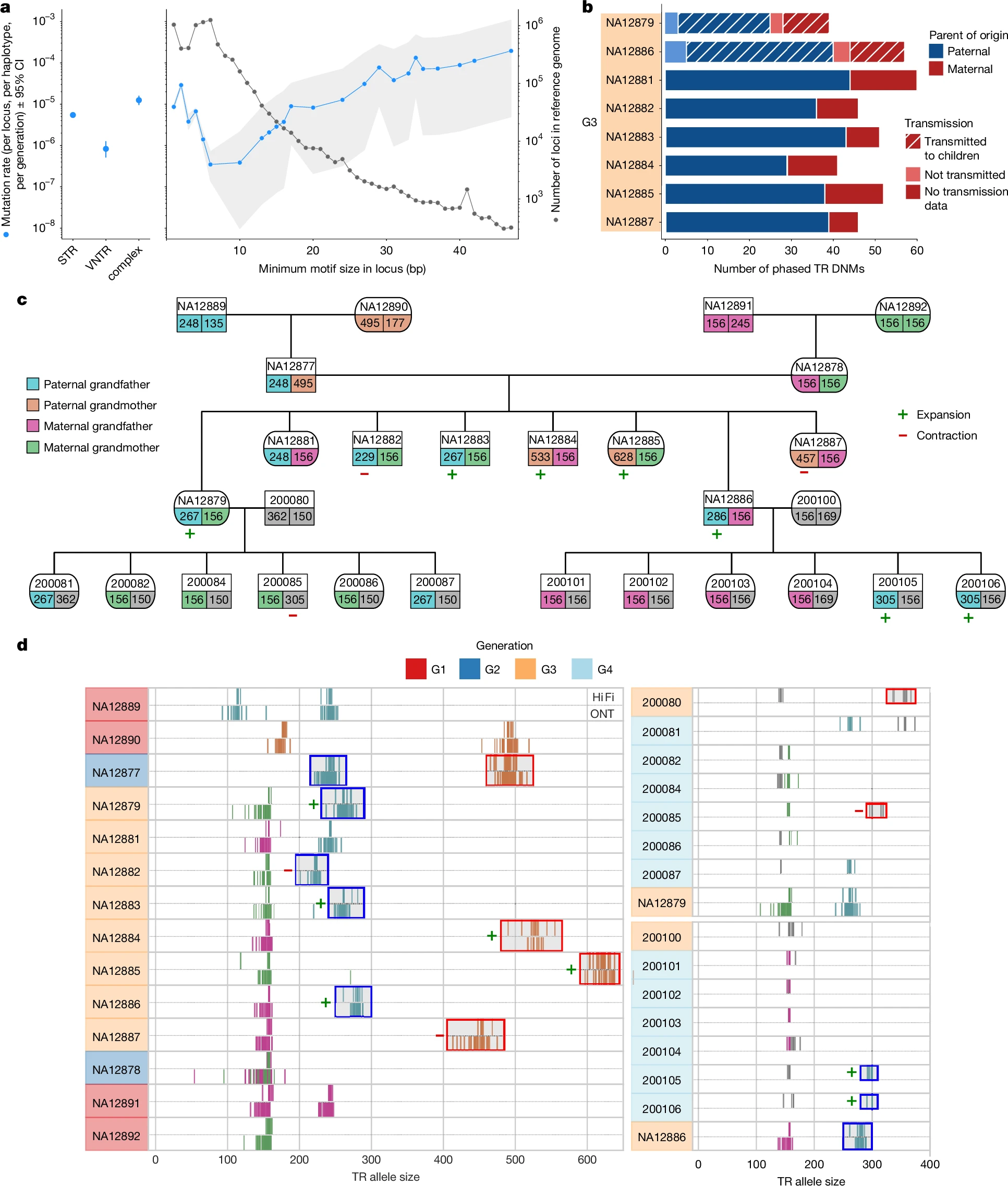
Human de novo mutation rates from a four-generation pedigree reference
Nature
·
23 Apr 2025
·
doi:10.1038/s41586-025-08922-2

STRchive: a dynamic resource detailing population-level and locus-specific insights at tandem repeat disease loci
Genome Medicine
·
26 Mar 2025
·
doi:10.1186/s13073-025-01454-4
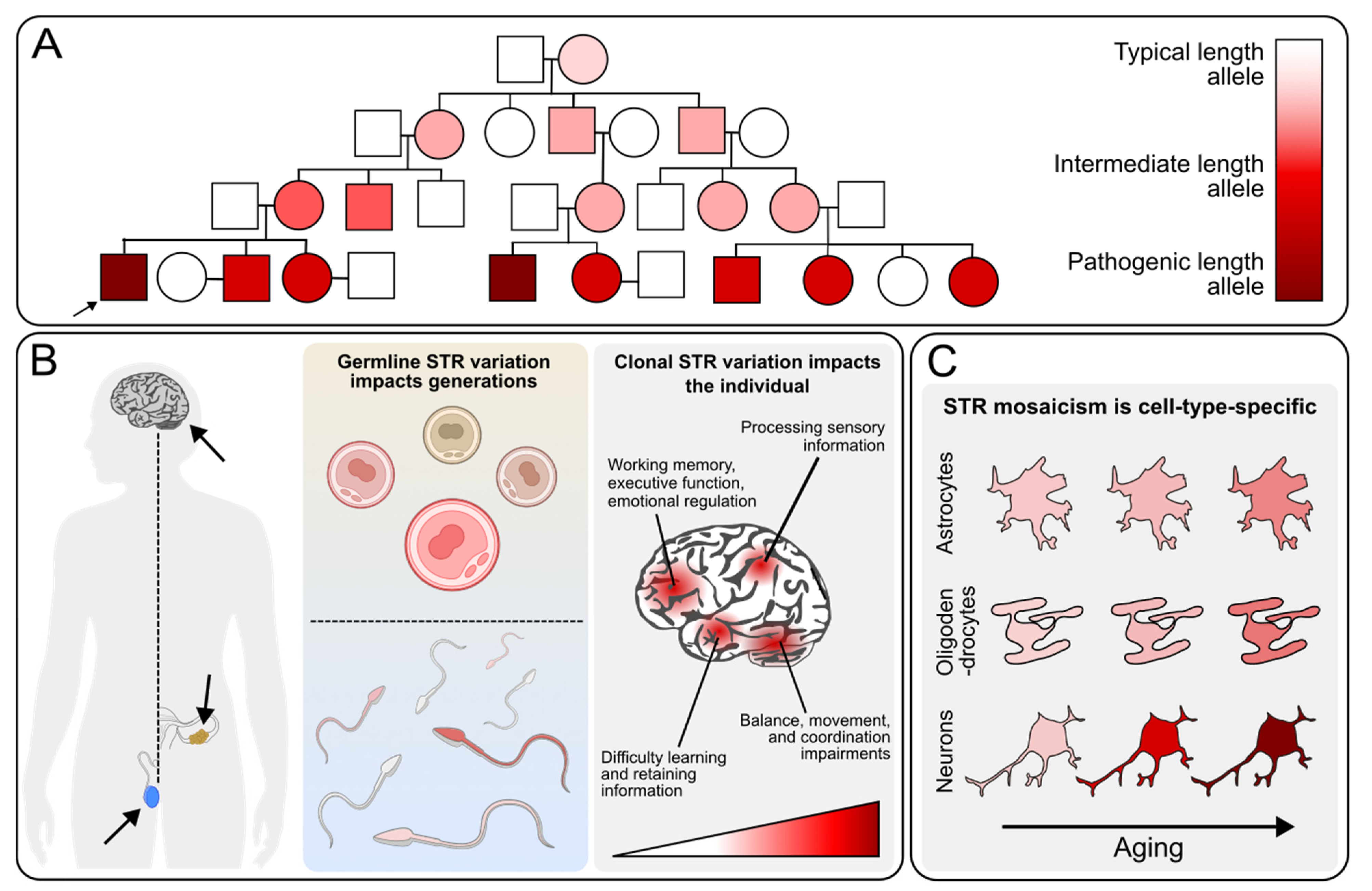
Mosaicism in Short Tandem Repeat Disorders: A Clinical Perspective
Genes
·
13 Feb 2025
·
doi:10.3390/genes16020216
2024

Polymorphic tandem repeats influence cell type-specific gene expression across the human immune landscape
Cold Spring Harbor Laboratory
·
04 Nov 2024
·
doi:10.1101/2024.11.02.621562

Defining a tandem repeat catalog and variation clusters for genome-wide analyses and population databases
Cold Spring Harbor Laboratory
·
05 Oct 2024
·
doi:10.1101/2024.10.04.615514

The Platinum Pedigree: A long-read benchmark for genetic variants
Cold Spring Harbor Laboratory
·
03 Oct 2024
·
doi:10.1101/2024.10.02.616333
High-coverage nanopore sequencing of samples from the 1000 Genomes Project to build a comprehensive catalog of human genetic variation
Genome Research
·
02 Oct 2024
·
doi:10.1101/gr.279273.124

A familial, telomere-to-telomere reference for human de novo mutation and recombination from a four-generation pedigree
Cold Spring Harbor Laboratory
·
05 Aug 2024
·
doi:10.1101/2024.08.05.606142

TRGT-denovo: accurate detection of de novo tandem repeat mutations
Cold Spring Harbor Laboratory
·
19 Jul 2024
·
doi:10.1101/2024.07.16.600745

Spotted with AuDHD: Seeing Yourself in a Mentor Who Sees You.
EdArXiv
·
01 Jun 2024
·
doi:10.35542/osf.io/pys86
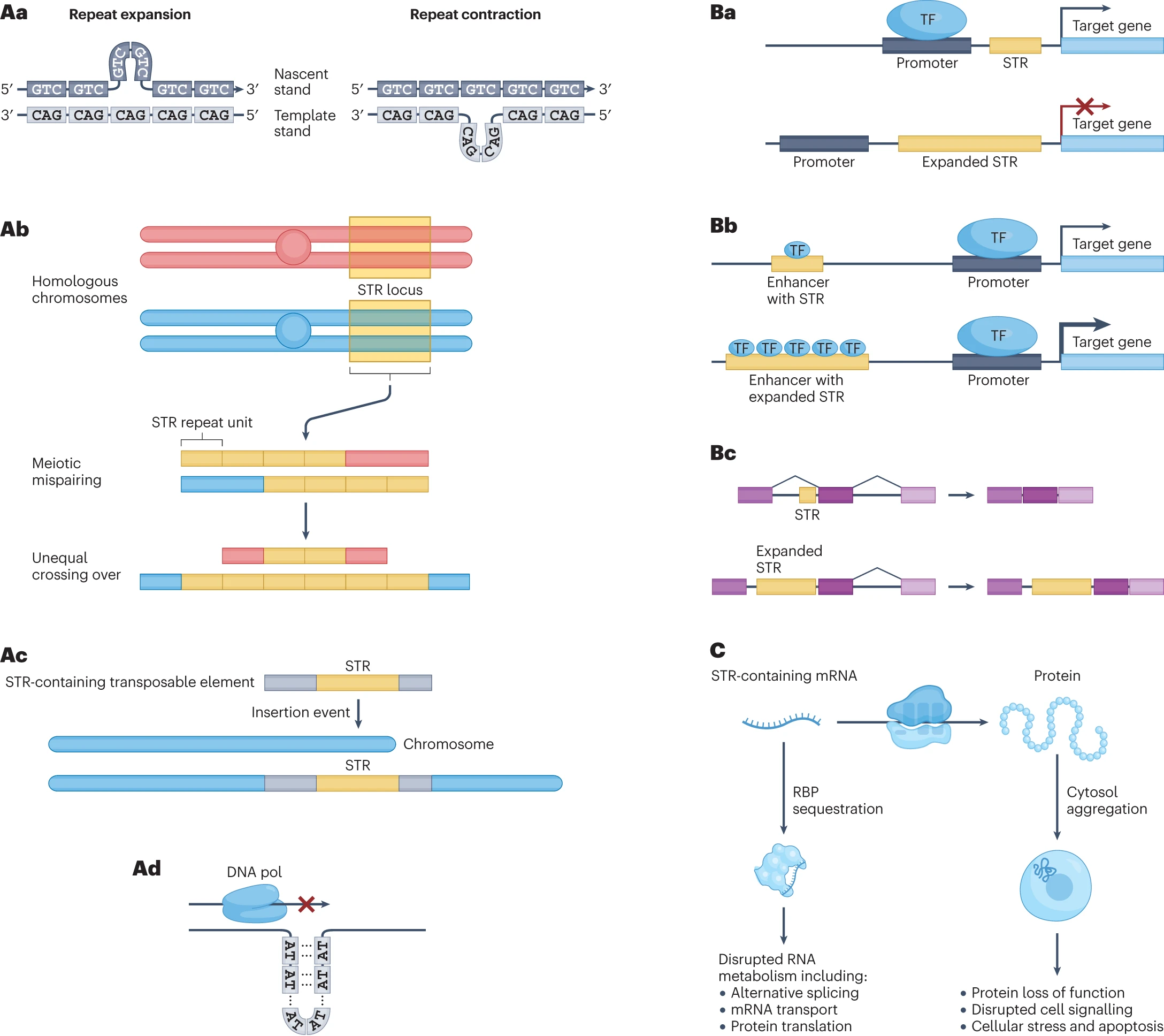
Sequencing and characterizing short tandem repeats in the human genome
Nature Reviews Genetics
·
16 Feb 2024
·
doi:10.1038/s41576-024-00692-3
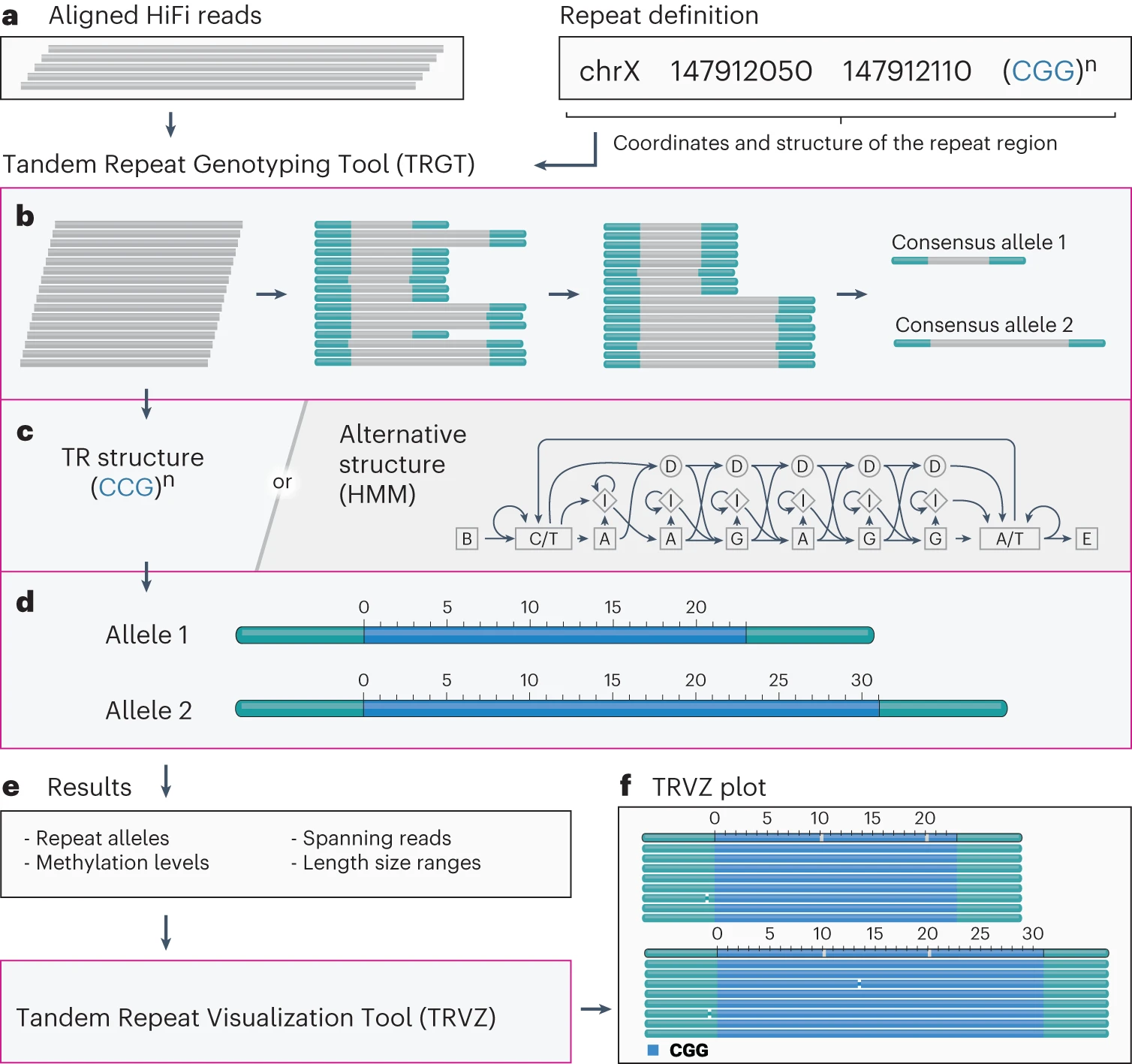
Characterization and visualization of tandem repeats at genome scale
Nature Biotechnology
·
02 Jan 2024
·
doi:10.1038/s41587-023-02057-3
2022
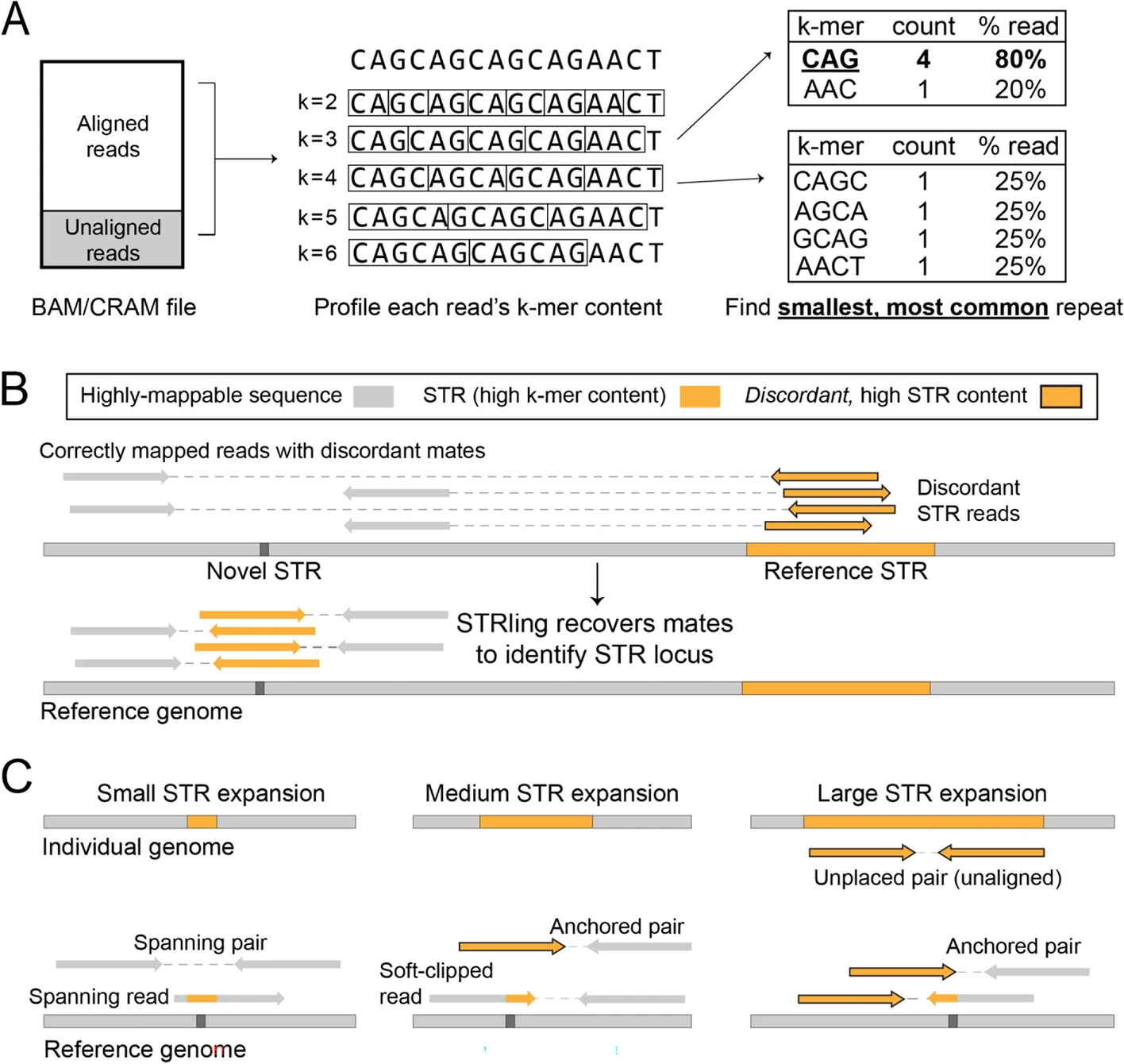
STRling: a k-mer counting approach that detects short tandem repeat expansions at known and novel loci
Genome Biology
·
14 Dec 2022
·
doi:10.1186/s13059-022-02826-4
2021
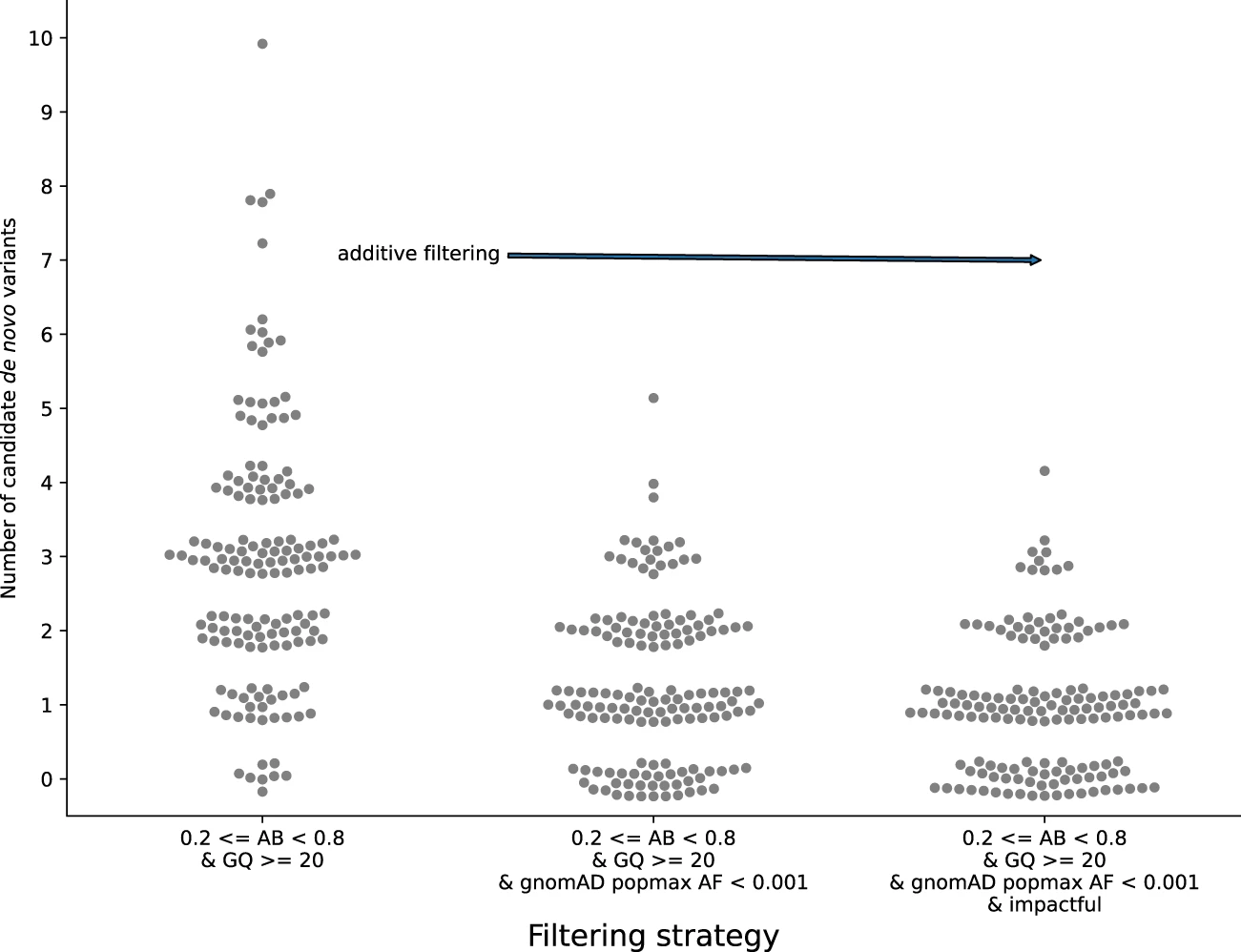
Effective variant filtering and expected candidate variant yield in studies of rare human disease
npj Genomic Medicine
·
15 Jul 2021
·
doi:10.1038/s41525-021-00227-3
2019
Bionitio: demonstrating and facilitating best practices for bioinformatics command-line software
GigaScience
·
01 Sep 2019
·
doi:10.1093/gigascience/giz109

Pooled-parent exome sequencing to prioritise de novo variants in genetic disease
Cold Spring Harbor Laboratory
·
07 Apr 2019
·
doi:10.1101/601740
2018
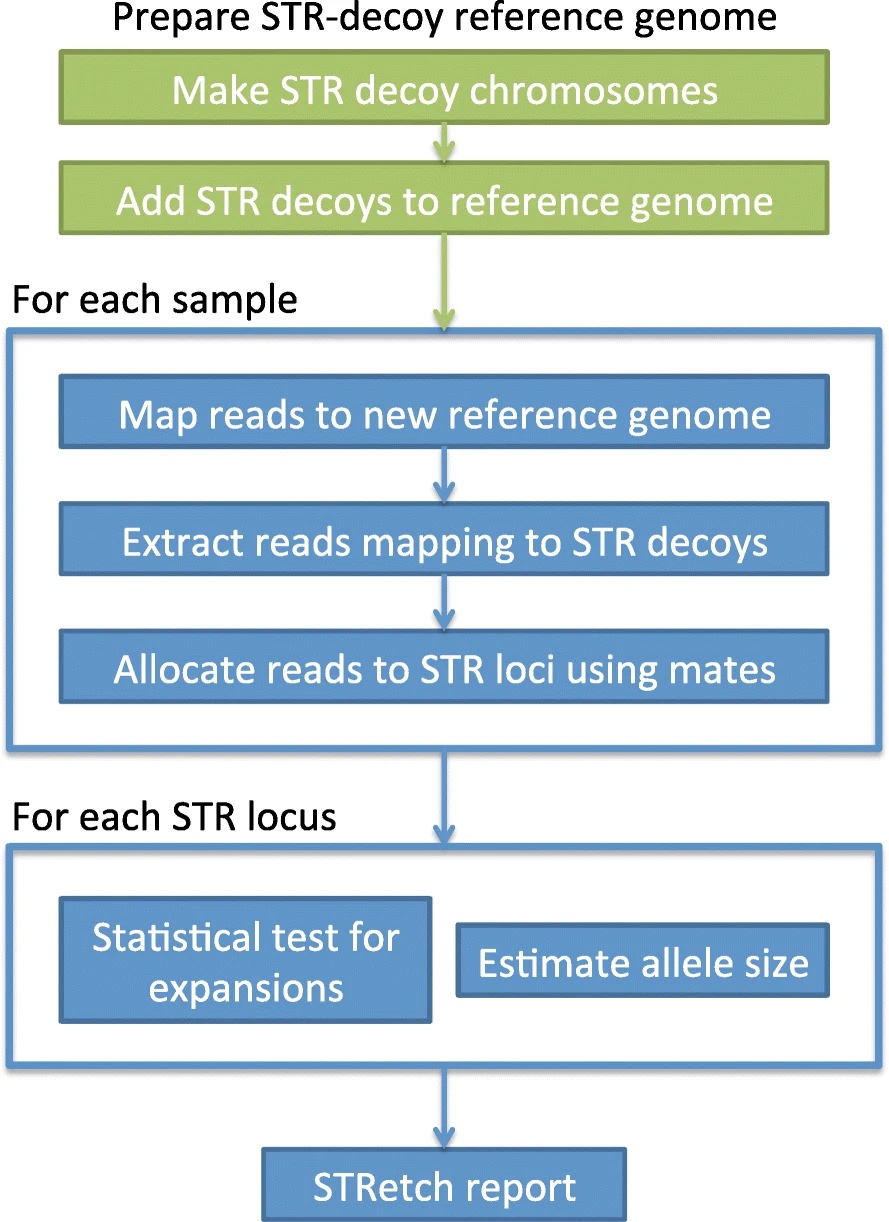
STRetch: detecting and discovering pathogenic short tandem repeat expansions
Genome Biology
·
21 Aug 2018
·
doi:10.1186/s13059-018-1505-2
2017

Elegant SciPy
O'Reilly Media, Inc.
·
08 Nov 2017
·
isbn:9781491922941

A clinically driven variant prioritization framework outperforms purely computational approaches for the diagnostic analysis of singleton WES data
European Journal of Human Genetics
·
23 Aug 2017
·
doi:10.1038/ejhg.2017.123
Melbourne Genomics variant prioritisation
2016
Ten Simple Rules for a Bioinformatics Journal Club
PLOS Computational Biology
·
28 Jan 2016
·
doi:10.1371/journal.pcbi.1004526
2015
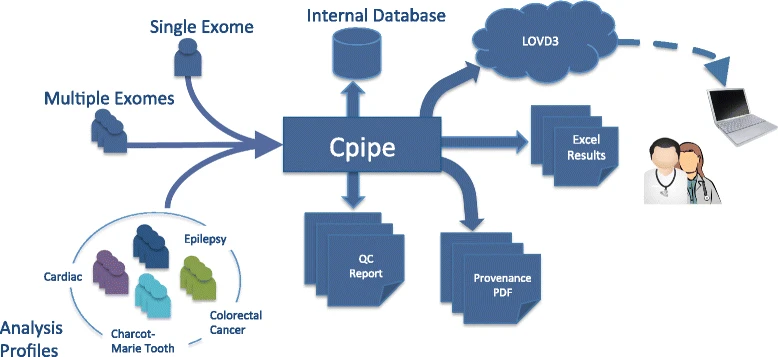
Cpipe: a shared variant detection pipeline designed for diagnostic settings
Genome Medicine
·
10 Jul 2015
·
doi:10.1186/s13073-015-0191-x
2014
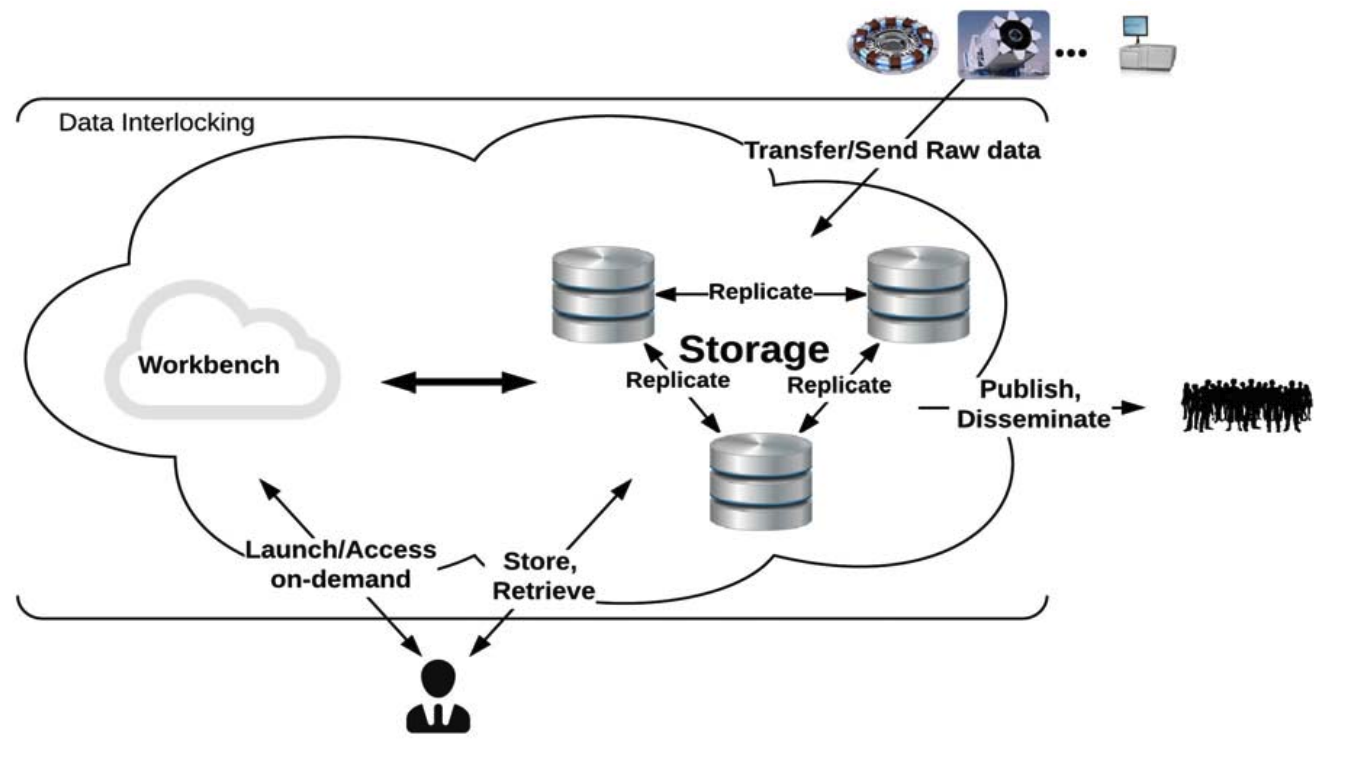
Data Interlocking: Coupling Analytics to the Data
2014 IEEE/ACM 7th International Conference on Utility and Cloud Computing
·
01 Dec 2014
·
doi:10.1109/ucc.2014.113
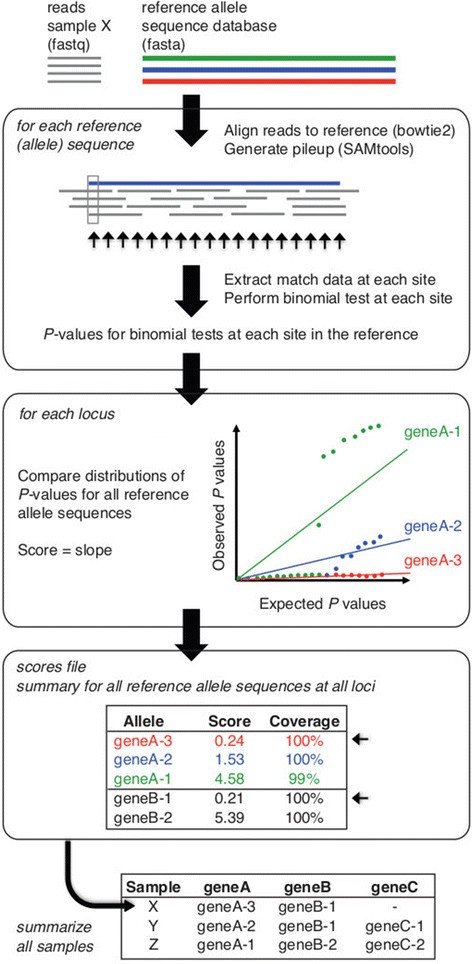
SRST2: Rapid genomic surveillance for public health and hospital microbiology labs
Genome Medicine
·
20 Nov 2014
·
doi:10.1186/s13073-014-0090-6
Ten Simple Rules for Writing a PLOS Ten Simple Rules Article
PLoS Computational Biology
·
23 Oct 2014
·
doi:10.1371/journal.pcbi.1003858
2012
Development of Transgenic Mice Containing an Introduced Stop Codon on the Human Methylmalonyl-CoA Mutase Locus
PLoS ONE
·
14 Sep 2012
·
doi:10.1371/journal.pone.0044974